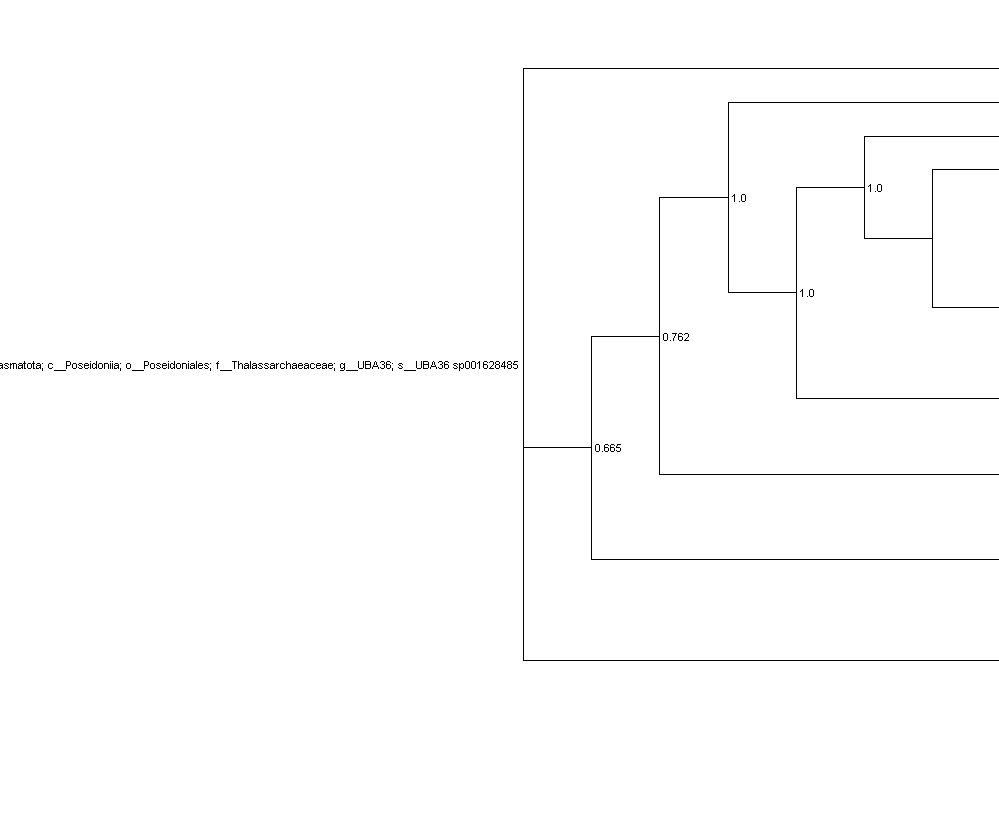Hi, I used the infer command to create a tree using the gtdbtk.ar53.user_msa.fasta.gz output from the align step of the de_novo_wf so I can visualize the archaea alone without the references using:
gtdbtk infer --msa_file gtdbtk.ar53.user_msa.fasta.gz --gamma --out_dir /mnt/f/PCARI-ASV/Analysis/Shotgun/gtdbtk_out/infer_archaea --cpus 20
However, when I decorate the tree above with the gtdbtk.ar53.summary.tsv output from the classify_wf as my classification file output, the tree comes out wrong.
gtdbtk decorate --input_tree /mnt/f/PCARI-ASV/Analysis/Shotgun/gtdbtk_out/infer_archaea/gtdbtk.unrooted.tree --output_tree /mnt/f/PCARI-ASV/Analysis/Shotgun/gtdbtk_out/decorate_archaea/decorate_archaea.tree --gtdbtk_classification_file /mnt/f/PCARI-ASV/Analysis/Shotgun/gtdbtk_out/classify_wf/gtdbtk.ar53.summary2.tsv
The first common label for the whole tree ends up being labelled as the species classification of some genomes:
It is consistent with the decorate_archaea.tree-taxonomy file where all genomes are assigned to d__Archaea; p__Thermoplasmatota; c__Poseidoniia; o__Poseidoniales; f__Thalassarchaeaceae; g__UBA36; s__UBA36 sp001628485
The decorate_archaea.tree-table also presents all 19 genomes as under the said lineage in contrast with the original classify_wf output where there are multiple different species level classifications.
The bacterial genomes I processed similarly seem to have proper decoration. Please let me know how I can fix this. Thanks!