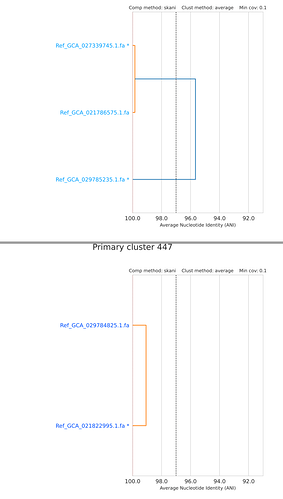
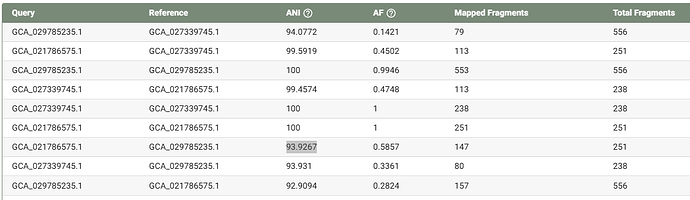
Recently, I performed an metagenomic analysis using AOA genomes downloaded from GTDB. However, I found some species shared ANI higher than 97%. In the ‘Q&A’ (GTDB - FAQ), the threshold to distinguish different species is 95% ANI and a maximum up to 97% ANI is also allowed (If I understand correctly). However, why these some species even shared ANI higher than 99% but still be assigned as different species? Here are some examples (not all):
These ANI plots were produced by dRep (97% threshold) based on all representive genomes of Nitrososphaeria. In other word, each genome represents a species in GTDB.