Hey,
First, would it be possible to reveal what combination of fields in the metadata file is used to determine whether a genome will be displayed as MAG or isolate in the online browser? This topic has been discussed in Assess if a genome is a MAG with *_metadata_r95.tsv files But I find the information incomplete.
As an example: GCF_016902615.1, s__Collinsella tanakei_A. How does GTDB derive that this is an isolate?
I looked up the ASM for this entry and further found that there they report
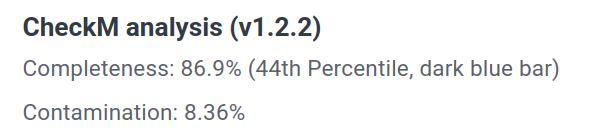
However in GTDB it says Completeness 100% and Contamination 0%.
Which is frankly quite confusing.
It would be great if you could shed some light on why that is, I am sure there is an explanation that I am not aware of.
Best,
Jogi
PS: I was gonna put the links in, but for new users there is a restriction on the number of links I can post.